Former Pikaard lab member, Dr. Keith Earley engineered a set of plant transformation vectors that allow for simple recombinational cloning of cDNA or genomic sequences using the Gateway cloning technology developed by Invitrogen (now part of ThermoFisher). We named these pEarleyGate vectors. The vectors were designed with transformation of Arabidopsis and other dicots in mind and are described in the following publication:
Earley, Keith, Jeremy R. Haag, Olga Pontes, Kristen Opper, Tom Juehne, Keming Song, and Craig S. Pikaard (2006). Gateway-compatible vectors for plant functional genomics and proteomics. The Plant J. 45:616-629.
The pEarleyGate vectors were all built by insertion of a Gateway cloning cassette, flanked by desired functional elements, into plasmid pFGC5491, a binary vector for Agrobacterium tumefaciens-mediated plant transformation that was built as part of the ChromDB (chromatin database) consortium (see Gendler et al., 2008, Nucleic Acids Research doi: 10.1093/nar/gkm768). pFGC5491, in turn, was engineered by modifying an open-source pCAMBIA vector backbone.
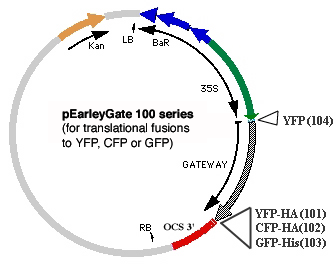
One set of pEarleyGate vectors (100 series) allows for rapid recombinational cloning of cDNAs, previously captured in Invitrogen pENTR vectors, to produce C-terminal fusions of encoded proteins in-frame with GFP, YFP or CFP or to produce N-terminal fusions to YFP. These vectors are useful for cell localization studies and use an enhanced Cauliflower Mosaic Virus (CaMV) 35S promoter to drive expression.
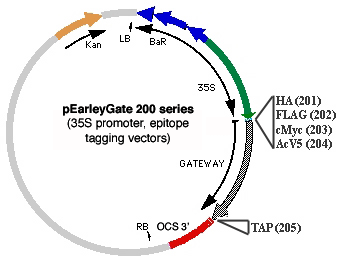
Another set of pEarleyGate vectors (200 series) allows for encoded proteins to be fused to HA, Myc, AcV5, or FLAG epitope tags, or to a tandem affinity peptide (TAP) tag, the latter consisting of a calmodulin binding peptide and a 2X Protein A peptide (which will bind to IgG resin) separated by a TEV protease cleavage site (Rigaut et al, 1999, Nature Biotechnology 17: 1030-1032). 200-series vectors also make use of a CaMV 35S promoter to drive expression.
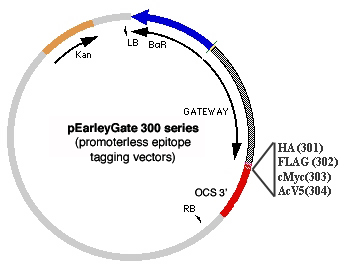
A third set of vectors (300 series) allows for expression of genes from a promoter of choice rather than from the enhanced 35S promoter. We find the 300 series vectors to be particularly useful for adding C-terminal tags to cloned genomic sequences that include the natural promoter, exons and introns, with the tag being added to the final exon in lieu of the natural stop codon.
All pEarleyGate vectors encode a kanamycin resistance cassette for plasmid selection in bacteria and a BAR gene cassette, encoding herbicide resistance, within the T-DNA for selection of transformed plants.
We made our pEarleyGate vectors freely available to the scientific community, without restrictions, by donating them to the Arabidopsis Biological Resource Center (ABRC), where they remain available and can be ordered online. A number of commercial companies are now selling pEarleyGate vectors, but there is no need to buy them! Please obtain them from ABRC and thereby support the stock center.
For a table describing the features of all pEarleyGate vectors, with links to maps, predicted sequences (based on parent vectors) and ordering information from the Arabidopsis stock center, please click here.
For notes on cloning sequences of interest into pEarleyGate vectors, please click here.


